
Chongzhi Zang Lab
Software
Transcriptional regulator analysis
 |
BART (Binding Analysis for Regulation of Transcription), a bioinformatics tool for inferring functional transcription factors (TFs) that bind at genomic cis-regulatory regions to regulate gene expression in the human or mouse genomes, given a query gene set or a ChIP-seq dataset as input. [Package] [Publication] |
 |
BART3D (BART for 3D genome data), a bioinformatics tool for inferring transcriptional regulators (TRs, including transcription factors and chromatin regulators) associated with differential chromatin interactions from Hi-C type 3D genome data for human or mouse. [Package] [Publication] |
 |
BARTsc (BART for single cell data), a bioinformatics tool for inferring transcriptional regulators (TRs, including transcription factors and chromatin regulators) that are specifically active for each cell cluster from single-cell omics data, including scRNA-seq, scATAC-seq, and single-cell multiome (ATAC+RNA) profiles. [Package (beta)] [Abstract] |
 |
MARGE (Model-based Analysis of Regulation of Gene Expression), a comprehensive computational method for inference of cis-regulation of gene expression leveraging public H3K27ac genomic profiles in human or mouse. [Package] [Publication] |
High-throughput omics bias correction and data integration
 |
PATTY (Pretrained Analyzer for Tn5 Transposase Yielded bias), a machine learning-based computational method for correcting open chromatin bias in bulk and single-cell CUT&Tag data. [Package (Beta)] [Preprint] |
 |
SELMA (Simplex Encoded Linear Model for Accessible chromatin), a computational model for estimation and correction of intrinsic enzymatic cleavage bias in bulk and single-cell chromatin accessibility profiling (DNase-seq and ATAC-seq) data. [Package] [Publication] |
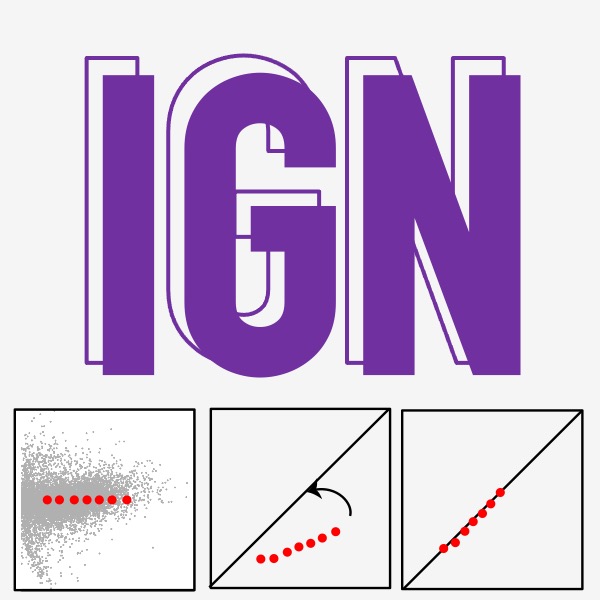 |
IGN (Invariable Gene Normalization), a computational method for chromatin profiling data normalization based on a set of unchanged genes. [Package] [Publication] |
 |
MANCIE (Matrix Analysis and Normalization by Concordant Information Enhancement), a computational method for high-dimensional genomic data integration. [Package] [Publication] |
Image-based chromatin profile analysis
 |
DORY (Differential chromatin tracing analysis), a computational method for identifying differential chromatin region pairs from image-based chromatin tracing data. [Package (Beta)] [Preprint] |
ChIP-seq signal detection
 |
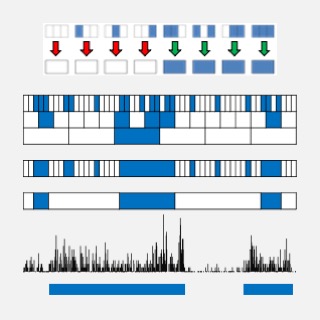
SICER (Spatial-clustering Identification of ChIP-Enriched Regions), a ChIP-Seq data analysis method. [Package (v1.1)] [Publication] |
 |
RECOGNICER (Recursive coarse-graining identification for ChIP-seq enriched regions), a ChIP-seq ultra-broad peak calling method. [Publication] [Source code] [Package built in SICER2] |
Resources
 |
BART Cancer, a database resource for computationally predicted transcriptional regulator activities in 15 human cancers from The Cancer Genome Atlas (TCGA). [Web resource] [Publication] |
Last modified: February 20, 2026